Can humans interbreed with other species?
No, don’t be ridiculous
What a disgusting suggestion! Uck!! A criminal conviction for bestiality will earn you a thumping prison-sentence and an odium that will shrink your social life. Anycase, have you forgotten the text-book definition of a species? That’s right; key to the concept are that populations must be reproductively isolated. So no inter-specific hanky-panky, thank you very much. It is just as well animals don’t read the text-books because in reality species do interbreed (and you should see the plants …). Such hybridization occasionally opens new evolutionary doors, but more often the outcome of such miscegenation is sterility. And what about us, Homo sapiens? No question that we descended from other species, but now alone we proudly stand. Would we ever stoop to interbreeding? Perish the thought!
Yes, of course
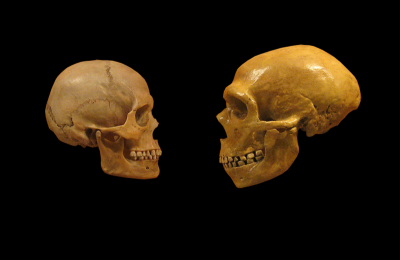
Our genomes tell a very different story. We may be alone, but a little beyond historical memory we had close companions in the form of the Neanderthals and the more recently discovered Denisovans. Skeletons of the former abound and reveal a powerfully built customer. Given Denisovans are known from only a couple of teeth and a tiny finger bone, reconstructing their original appearance might seem a tall order. Let’s ask Holmes: “Observe, Watson, mere scraps, but the tooth, is it not somewhat archaic? Summon one of my Irregulars, I want this finger-bone to be taken poste-haste to that ingenious DNA lab in Leipzig …”. Ingenious indeed, because not withstanding almost invariably massive microbial contamination, original DNA is not only recovered but it confirms that although Denisovans, humans and Neanderthals are genomically near-identical there real differences in the DNA and they yield extraordinary insights. First, Denisovans and Neanderthals are the closer cousins (separating about 550,000 years ago), whilst we and Neanderthals went our separate ways perhaps a quarter of million years later. Did I say “separate”? Not quite. Lurking in our genomes are chunks of Denisovan and Neanderthal DNA and they didn’t get there by sneezing. The only explanation is interbreeding, in the trade this transfer is known as introgression. The story, however, is a bit more complicated. Writing as a Eurasian roughly 2% of my DNA is Neanderthal. Amongst Asians this figure is slightly higher, but if we look at sub-Saharan Africa then with the curious exception of the Maasai only a trace of this imported DNA can be found. So did they escape introgression? Not at all; sub-Saharan people carry a tell-tale signature of a separate encounter with another archaic hominin, and perhaps as recently as 35,000 years ago. The Denisovan story is if anything more intriguing. In the inhabitants of Papua New Guinea about 6% of the DNA is of Denisovan origin and appreciable amounts are also found in surrounding populations. Whilst these genomic footprints provide unequivocal evidence of interbreeding, successful hybridizations were still extremely rare. One estimate suggests one successful mating roughly every eighty generations will suffice to ensure the transfer of Neanderthal or Denisovan DNA.
It all depends on the question
So does this introgressed DNA make us any less human? Not at all! First of all humans readily interbreed. Whether or not you are the proud carrier of Neanderthal or Denisovan DNA makes no difference at all. We are one species. Note also that the original quantities of imported DNA were probably greater, but some of the visitor DNA has been shown to the door. It is no coincidence that in this respect the X-chromosome is notably “clean”; on this sex-chromosome introgressed DNA might do real damage. Elsewhere, however, there might be real advantages, although deciding exactly how is more tricky. First, by no means all living human populations have the same profiles of introgression so any benefits cannot be universal. Second, genomic history is complex; in some cases the DNA may actually derive from an older common ancestor. This is important because features such as language might significantly predate the emergence of Neanderthals, Denisovans and ourselves. Third, genes are usually multifunctional and it is risky to say gene A does only function X. Even so there are some interesting links. Most striking, perhaps, is the clear connection between the remarkable adaptability of Tibetans to high altitude-life and what is clearly a Denisovan import. To be sure other links are more tenuous, but they may include skin colour (and hence UV protection or vitamin D synthesis), skeletal structure, the immune system and cognitive capacities.
Details of when, where and how often these hominin species met are sketchy, but the introgressed DNA we carry is mute testimony to a far more complex history of distributions, contacts and migrations than was once imagined. And as the unexpected discovery of the “hobbits” on Flores (at conferences they are called Homo floresiensis) should remind us, maybe there were still other early hominins roving the planet at the same time, yet to be discovered? Remember that currently the Denisovans are known from only a single Siberian cave. To explain, however, where the remnants of their DNA are now found can only be explained if originally they had a huge geographical range, rivalling that of the Neanderthals. The Denisovans are yet to tell their full story, but in the case of the Neanderthals a persistent idea is that despite their success ultimately their extinctions were the result of coming second-best, runners-up against the smarter us. Maybe so, but evidence from diets, cooking, hunting, clothing, technology and most importantly cultural and symbolic activities suggest that it could have as easily been a Neanderthal writing these lines. Maybe their fatal flaw was tiny populations that were too vulnerable. Even so, Neanderthals possessed speech … and imagination. Forever gone, now they haunt our imaginations. It is an early summer day near the River Jordan and mist floats across the Palaeolithic meadow. Was it an accident or already arranged? Either way Neanderthal and human, male and female meet. Boy meets girl as they say, but this time across the species boundary. 50,000 years later we still carry a clear genetic memory of this fleeting meeting.
Text copyright © 2015 Simon Conway Morris. All rights reserved.
Further reading
Abi-Rached, L. et al. (2011) The shaping of modern immune systems by multiregional admixture with archaic humans. Science 334, 89-94.
Ding, Q-L. et al. (2014) Neanderthal introgression at chromosome 3p21.31 was under positive natural selection in east Asians. Molecular Biology and Evolution 31, 689-695.
Fu, Q-M. et al. (2015) An early modern human from Romania with a recent Neanderthal ancestor. Nature 524, 216-219.
Gokhman, D. et al. (2014) Reconstructing the DNA methylation maps of the Neanderthal and the Denisovan. Science 344, 523-527.
Hardy, B.L. et al. (2013) Impossible Neanderthals? Making string, throwing projectiles and catching small game during Marine Isotope Stage 4 (Abri du Maras, France). Quaternary Science Reviews 82, 23-40.
Hawks, J. (2013) Significance of Neandertal and Denisovan genomes in human evolution. Annual Review of Anthropology 42, 433-449.
Huerta-Sánchez, E. et al. (2014) Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194-197.
Lachance, J. et al. (2012) Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse hunter-gatherers. Cell 150, 457-469.
Mendez, F.L. et al. (2012) A haplotype at STAT2 introgressed from Neanderthals and serves as a candidate of positive selection in Papua New Guinea. American Journal of Human Genetics 91, 265-274.
Meyer, M. et al. (2012) A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222-226.
Neves, A.G.M. and Serva, M. (2012) Extremely rare interbreeding events can explain Neanderthal DNA in living humans. PLoS ONE 7, e47076.
Overmann, K.A. and Coolidge, F.L. (2013) Human species and mating systems: Neandertal-Homo sapiens reproductive isolation and the archaeological and fossil records. Journal of Anthropological Sciences 91, 91-110.
Prüfer, K. et al. (2014) The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43-49.
Sankararaman, S. et al. (2012) The date of interbreeding between Neandertals and modern humans. PLoS Genetics 8, e1002947.
Sankararaman, S. et al. (2014) The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507, 354-357.
Wall, J.D. et al. (2013) Higher levels of Neanderthal ancestry in East Asians than in Europeans. Genetics 194, 199-209.